- Whole transcriptome coverage with probes specific to protein coding mRNA sequences
- Compatible with common sample types such as formalin-fixed paraffin embedded (FFPE) or fresh frozen (FF) tissue and across all human tissues types
- Superior sensitivity to detect 1000s of unique human genes in <50 μm regions
- Robust performance across sample types including FFPE with high concordance with RNA-seq and RNAscope™
- Map single cell RNA-seq populations to their tissue location
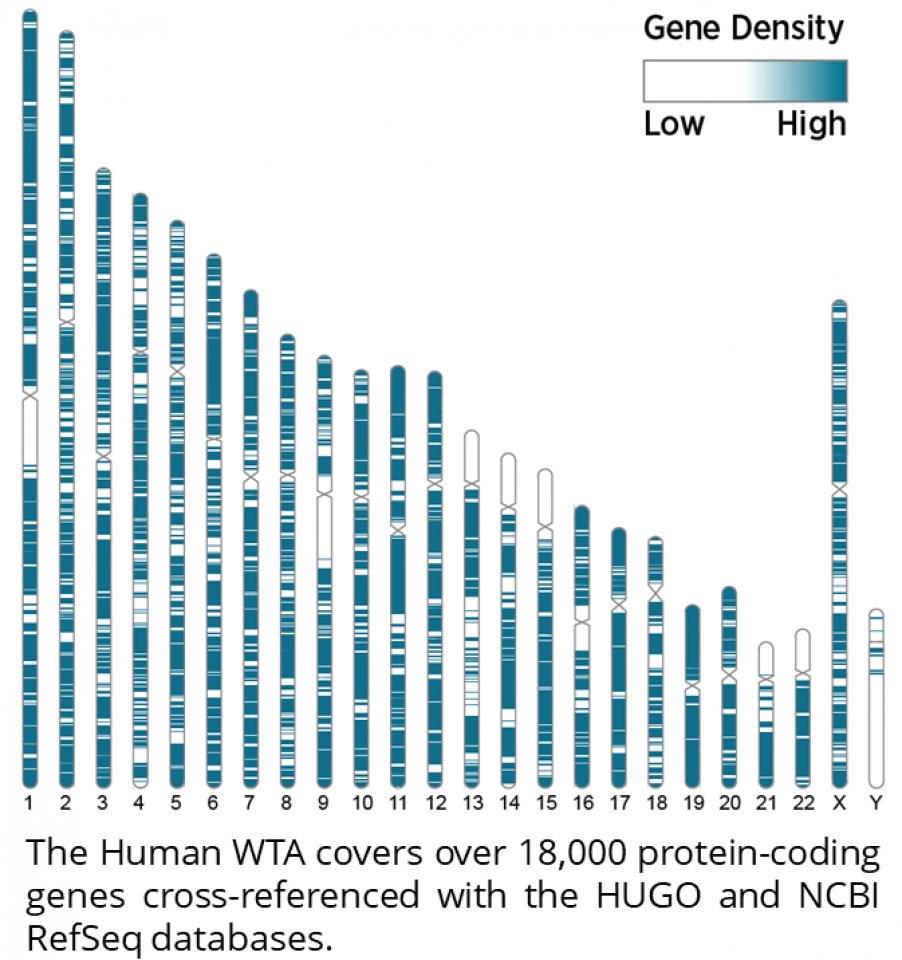
The GeoMx Whole Transcriptome Atlas delivers the maximum amount of sensitivity and confidence in each transcript through its unique probe architecture. The WTA profiles over 18,000 protein-coding human genes based on the human gene nomenclature committee (HUGO1) database cross-referenced with available mRNA sequences in the National Center for Biotechnology’s Information (NCBI) RefSeq database.
The WTA then allows to explore pathways across the whole transcriptome in defined regions of interest.
Learn about out Digitial Spatial Profiling (DSP) services
- Extensive coverage of the immune response, tissue microenvironment, tumor biology, and genes from clinically relevant genes sets such as tumor inflammation
- Spatial measurement of single cell signatures with high sensitivity and dynamic range
- Inclusion of genes for the Tumor Inflammation Signature, PAM50 and other clinical signatures
- Over 100 pathways to explore all aspects of cancer and tumor biology
Learn about out spatial transcriptomics services
Comprehensively Annotated Pathways in the CTA
| Adaptive Immunity | |
Cell Function | |
Signaling Pathways |
| T cells B cells | Apoptosis | AMPK Signaling | ||
| TCR & BCR Signaling | Autophagy | Androgen Signaling | ||
| Cancer Antigens | Cell Adhesion & Motility | EGFR Signaling | ||
| MHC Class I & II Antigen Presentation | Cell Cycle | ERBB2 Signaling | ||
| T-cell Checkpoints | Cilium Assembly | Estrogen Signaling | ||
| TH1, TH2, TH9, Th17, and Treg Differentiation | Differentiation | FGFR Signaling | ||
| DNA Damage Repair | FoxO Signaling | |||
| Innate Immunity | EMT | GPCR Signaling | ||
| Complement System | Endocytosis | Hedgehog Signaling | ||
| Dendritic Cells | Epigenetic Modification | HIF1 Signaling | ||
| DNA & RNA Sensing | Immortality & Stemness | Insulin Signaling | ||
| Glycan Sensing | Ion Transport | JAK-STAT Signaling | ||
| Host Defense Peptides | Lysosome | MAPK Signaling | ||
| Inflammasomes | Oxidative Stress | MET Signaling | ||
| Myeloid Inflammation | Phagocytosis | mTOR Signaling | ||
| Neutrophil Degranulation | Proteotoxic Stress | Myc | ||
| NK Activity | RNA Processing | NO Signaling | ||
| NLR Signaling | Senescence | Notch Signaling | ||
| RAGE Signaling | p53 Signaling | |||
| TLR Signaling | Metabolism | PDGF Signaling | ||
| Amino Acid Synthesis & Transport | PI3K-Akt Signaling | |||
| Immune Response | Arginine & Glutamine Metabolism | PPAR Signaling | ||
| Chemokine Signaling | Fatty Acid Oxidation & Synthesis | Purinergic Signaling | ||
| Cytotoxicity | Glycolysis & Glucose Transport | Retinoic Acid Signaling | ||
| IL-1, IL-2, IL-6 & IL-17 Signaling | Glycosylation | TGF-beta Signaling | ||
| Immune Exhaustion | IDH1/2 | VEGF Signaling | ||
| Interferon Response Genes | Lipid Metabolism | Wnt Signaling | ||
| Lymphocyte Regulation & Trafficking | Mitochondrial Metabolism / TCA | |||
| NF-kB Signaling | Nucleotide Synthesis | Physiology & Disease | ||
| Other Interleukin Signaling | Pentose Phosphate | Angiotensin System | ||
| Prostaglandin Inflammation | Tryptophan & Kynurenine Metabolism | Circadian Clock | ||
| TNF Signaling | Vitamin & Cofactor Metabolism | Drug Resistance | ||
| Type I, II, & III Interferon Signaling | |
Glioma | ||
| |
Leukemia | |||
| Matrix Remodeling and Metastasis | ||||
| Melanoma | ||||
| Neuroendocrine Function | ||||
| Prostate Cancer |