The development of immune checkpoint blockers (ICB) has considerably changed the therapeutic armamentarium for the management of cancer. Blocking the PD-1/PD-L1 axis interaction has demonstrated remarkable anti-cancer activity and has led to approval of anti-PD-1/PD-L1 drugs in several solid tumors. However, most patients receiving PD(L)-1 blockers do not derive clinical benefit. Therefore, there is a crucial need to decipher mechanisms underlying sensitivity / resistance to cancer immunotherapies which can ultimately lead to the identification of novel therapeutic targets.
Through a collaborative work (with Institut Bergonié) using the GeoMx-DSP technology platform (NanostringTM) and the GeoMx® Immune Pathways Panel, and aiming at delineating biological pathways involved in resistance / sensitivity of NSCLC to anti-PD-(L)1-based immunotherapies, we investigated the spatial transcriptomic profile of tumor samples collected before treatment onset of responder (and non-responder patients. Tumor- and Stroma-specific gene expression was analyzed through the application of morphology markers including PanCytoKeratin (PanCK) for tumor cells and CD45 for immune cells.
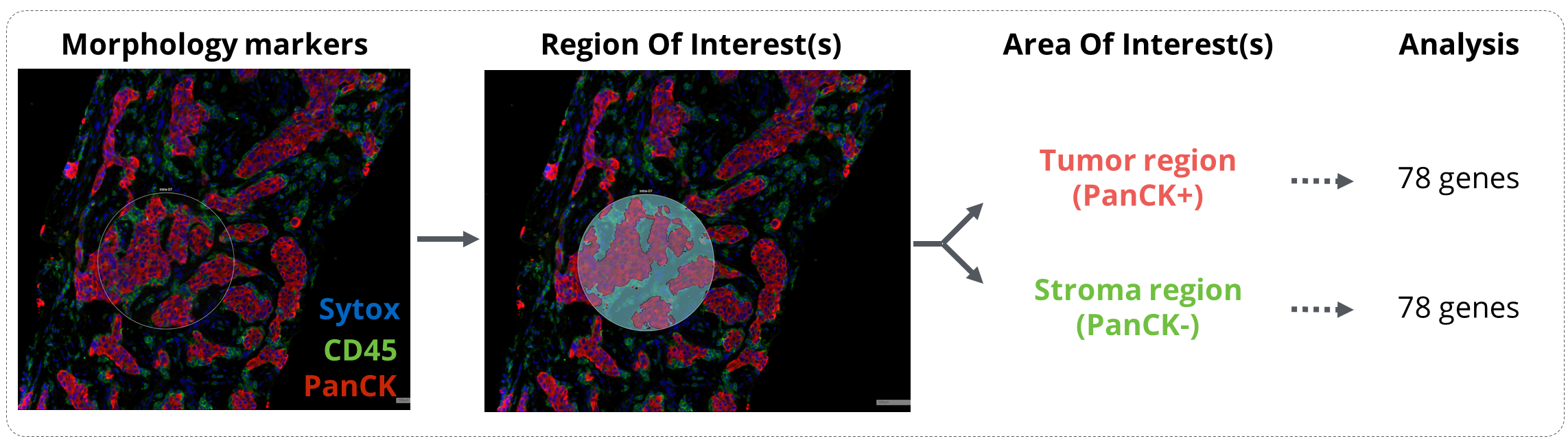
Figure 1. GeoMx workflow analysis. Tissues were incubated with the GeoMx® Immune Pathways Panel probes and stained with the morphology markers CD45 and PanCytokeratin (PanCK). Regions of interest were drawn in mixed tumor/stroma areas and were then segmented in Tumor (PanCK) and Stroma (PanCK-) compartments. Expression of 78 protein-coding genes related to tumor microenvironment profiling were finally analyzed on the Nanostring nCounter platform.
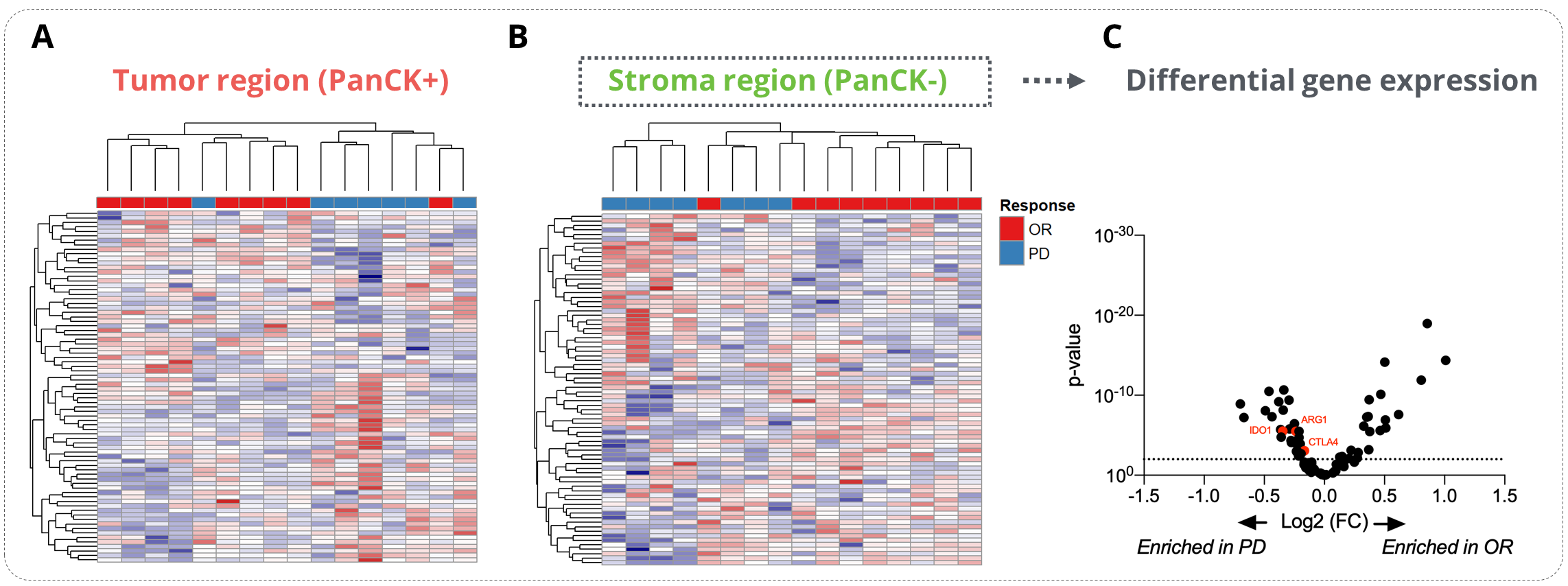
Unsupervised data clustering evidenced a strong stratification of responder and non-responder patients with data collected from the stroma compartment. Using this dataset, we performed a differential gene expression analysis between responders and patients with a progressive disease (PD). This analysis allowed to reveal key determinants of response to ICB in NSCLC patients whose some genes involved in immune escape, e.g. Ctla4, Arg1, Ido1, were enriched in PD patients.
Figure 2. Analysis of the determinants of the ICI response. Sixteen tumor samples from NSCLC patients (8 responders – OR and 8 non-responders – PD) were analyzed on the GeoMx® DSP using the GeoMx® Immune pathways panel (11-12 ROIs per patients). (A-B) Heatmap representations of the tumor (A) and stroma (B) specific gene expression. (C) Differential gene expression analysis of the stroma-specific gene expression from responders and non-responders patients . OR stands for Objective Response and PD for Progressive Disease.
Besides revealing known and novel markers underlying sensitivity to ICB in NSCLC patients, this novel dataset illustrates the key benefit of the spatial transcriptomic approach that can be conducted with a limited and target panel (e.g. immune pathway) or with a more comprehensive approach using Cancer or Whole Transcriptome Atlas panels (1800 and 18000 genes, respectively).
Learn more about our platform in spatial transcriptomics