In a groundbreaking study led by Prof. Antoine Italiano from the Bergonié & Gustave Roussy Comprehensive Cancer Centers, 42 patients with advanced gastroenteropancreatic neuroendocrine neoplasms were treated with a combination of anti-PD-1/PD-L1 immunotherapy and the angiogenesis inhibitor regorafenib. The combination therapy resulted in increased response rates and improved survival outcomes compared to PD-1/PD-L1 immunotherapy alone. However, a significant portion of patients did not respond to this therapeutic regimen.
To uncover the mechanisms of resistance and identify potential predictive biomarkers, Explicyte developed a custom immunohistofluorescence (IHF) panel. Digital pathology revealed an overall increase in CD8+ T cell infiltration. In tumors from non-responding patients, we observed a higher proportion of PD1+ / IDO1+ neoplastic cells, suggesting the presence of an immunosuppressive environment mediated by tryptophan metabolism along the kynurenine pathway. This hypothesis was further supported by quantifying the Kynurenine/Tryptophan ratio, which was associated with IDO1 expression in tumor tissues and correlated with poorer clinical outcomes. Additionally, using Olink’s immuno-oncology and inflammation panels, we identified an enrichment of immune related proteins in the plasma of patients with high IDO1 activity. Soluble PD-L1, TNF receptor family members (OX40, CD40, 4-1BB), and the immunosuppressive cytokine interleukin-10 were associated with unfavorable outcomes.
Altogether, this translational study suggests shows that the combination of regorafenib plus avelumab is interesting for patients with highly pre-treated GEP NETs, and suggests that adding IDO1 inhibitors to the combination of anti-PD1/PDL1 therapy and anti-angiogenic agents could be a promising strategy to enhance patient responses.
To uncover the mechanisms of resistance and identify potential predictive biomarkers, Explicyte developed a custom immunohistofluorescence (IHF) panel. Digital pathology revealed an overall increase in CD8+ T cell infiltration. In tumors from non-responding patients, we observed a higher proportion of PD1+ / IDO1+ neoplastic cells, suggesting the presence of an immunosuppressive environment mediated by tryptophan metabolism along the kynurenine pathway. This hypothesis was further supported by quantifying the Kynurenine/Tryptophan ratio, which was associated with IDO1 expression in tumor tissues and correlated with poorer clinical outcomes. Additionally, using Olink’s immuno-oncology and inflammation panels, we identified an enrichment of immune related proteins in the plasma of patients with high IDO1 activity. Soluble PD-L1, TNF receptor family members (OX40, CD40, 4-1BB), and the immunosuppressive cytokine interleukin-10 were associated with unfavorable outcomes.
Altogether, this translational study suggests shows that the combination of regorafenib plus avelumab is interesting for patients with highly pre-treated GEP NETs, and suggests that adding IDO1 inhibitors to the combination of anti-PD1/PDL1 therapy and anti-angiogenic agents could be a promising strategy to enhance patient responses.
Read article in Nature Cancer
Join us for an insightful session on March 27th, featuring the first authors behind three influential 2025 papers on tertiary lymphoid structures (TLS) in cancer:
Presenting a groundbreaking 2025 Nature paper, "IL-33-activated ILC2s induce tertiary lymphoid structures in pancreatic cancer". Dr. Amisaki’s work deepens our understanding of TLS biology and paves the way for innovative therapeutic strategies for transforming “cold” tumors into immunologically active environments.
Sharing findings from his PhD research, conducted at Explicyte, Dr. Peyaud will discuss 2 publications:
Immunologist Alban Bessede, PhD, CEO of Explicyte, will kick off the session. Dr Bessede has been working on TLS since 2020, co-authoring foundational papers in Nature Cancer and Nature Medicine on TLS & immune-checkpoint inhibitors, and is now developing accredited pathology services for detecting and scoring TLS in tumor specimens to support precision oncology initiatives.
Masataka Amisaki, MD, PhD, MSKCC (USA)
Presenting a groundbreaking 2025 Nature paper, "IL-33-activated ILC2s induce tertiary lymphoid structures in pancreatic cancer". Dr. Amisaki’s work deepens our understanding of TLS biology and paves the way for innovative therapeutic strategies for transforming “cold” tumors into immunologically active environments.
Florent Peyraud, MD, PhD, Institut Bergonié (France)
Sharing findings from his PhD research, conducted at Explicyte, Dr. Peyaud will discuss 2 publications:
- A research paper in Cell Reports Medicine titled "Spatially resolved transcriptomics reveal the determinants of primary resistance to immunotherapy in NSCLC with mature tertiary lymphoid structures"
- A comprehensive review in Med. titled "Tertiary lymphoid structures and cancer immunotherapy: From bench to bedside"
Session Introduction
Immunologist Alban Bessede, PhD, CEO of Explicyte, will kick off the session. Dr Bessede has been working on TLS since 2020, co-authoring foundational papers in Nature Cancer and Nature Medicine on TLS & immune-checkpoint inhibitors, and is now developing accredited pathology services for detecting and scoring TLS in tumor specimens to support precision oncology initiatives.
Register for the webinar
This translational research paper highlights the work of Florent Peyraud, MD, conducted during his PhD thesis at Explicyte under the supervision of Prof. Antoine Italiano (Bergonié & Gustave Roussy Comprehensive Cancer Centers) and Alban Bessede, PhD (Explicyte). This project was made possible thanks to a collaboration with the imCORE network (Roche/Genentech), and samples from the BIP trial (NCT02534649). The study aims to uncover why certain patients with non-small cell lung cancer (NSCLC) who exhibit mature tertiary lymphoid structures (mTLS)—a phenotype typically associated with a favorable response to immune checkpoint inhibitors (ICI)—do not respond to anti-PD1/PD-L1 immunotherapy.
Presence of mature TLS & clinical outcome in NSCLC
A comprehensive pathological analysis of 509 NSCLC patients treated with immune checkpoint inhibitors (ICI) was performed, confirming that the presence of mTLS predicts a better outcome in terms of response rate, overall survival, and median progression-free survival, independently of PD-L1 and genomic features.
Spatial transcriptomics in responding & non-responding mTLS-positive NSCLC patients
Specimens from 6 mTLS-positive patients with extreme ICI response (3 responders, 3 non-responders) were analyzed using the GeoMx Whole Transcriptome Atlas. While gene expression profiles did not differ within the TLS and tumor compartments, non-responders exhibited an enrichment in fibroblasts in the stromal compartment, with a pronounced expression of TGFβ signaling and epithelial-mesenchymal transition (EMT) pathways.
Multiplex immunohistofluorescence (mIHF) panel focusing on cancer-associated fibroblasts (CAFs) 77 m-TLS positive patient samples were then analyzed by digital pathology for the detection of tumor cells, cytotoxic CD8+ T cells, and FAP+αSMA+ CAF and MYH11+αSMA+ CAFs. Analysis revealed a higher stromal density of both CAF subsets in non-responders, correlating with poor clinical outcome.
Bulk transcriptomics to analyze interactions between the immune infiltrates & CAF status
Gene expression profiling of 40 mTLS-positive lung tumor samples identified that:
Multiplex immunofluorescence panel focused on T cell exhaustion
Specimens of 64 mTLS positive NSCLC patients were then analyzed by digital pathology:
Altogether, these findings suggest that the presence of FAP+αSMA+ CAFs and MYH11+αSMA+ CAFs is associated with poor outcomes in mTLS-positive NSCLC patients undergoing ICI treatment. They could serve as biomarkers to stratify patients, and could constitute targets, in combination with strategies addressing T-cell exhaustion, to enhance ICI efficacy in mTLS-positive NSCLC patients.
Interested in the assessment of TLS maturity? Learn about our histopathology services for TLS detection & scoring.
Presence of mature TLS & clinical outcome in NSCLC
A comprehensive pathological analysis of 509 NSCLC patients treated with immune checkpoint inhibitors (ICI) was performed, confirming that the presence of mTLS predicts a better outcome in terms of response rate, overall survival, and median progression-free survival, independently of PD-L1 and genomic features.
Spatial transcriptomics in responding & non-responding mTLS-positive NSCLC patients
Specimens from 6 mTLS-positive patients with extreme ICI response (3 responders, 3 non-responders) were analyzed using the GeoMx Whole Transcriptome Atlas. While gene expression profiles did not differ within the TLS and tumor compartments, non-responders exhibited an enrichment in fibroblasts in the stromal compartment, with a pronounced expression of TGFβ signaling and epithelial-mesenchymal transition (EMT) pathways.
Multiplex immunohistofluorescence (mIHF) panel focusing on cancer-associated fibroblasts (CAFs) 77 m-TLS positive patient samples were then analyzed by digital pathology for the detection of tumor cells, cytotoxic CD8+ T cells, and FAP+αSMA+ CAF and MYH11+αSMA+ CAFs. Analysis revealed a higher stromal density of both CAF subsets in non-responders, correlating with poor clinical outcome.
Bulk transcriptomics to analyze interactions between the immune infiltrates & CAF status
Gene expression profiling of 40 mTLS-positive lung tumor samples identified that:
- FAP+αSMA+ CAF-High tumors were associated with inflammatory response and T-cell exhaustion, which may explain the limited efficacy of ICI.
- MYH11+αSMA+ CAF-High tumors were associated with an enrichment in regulatory T cell (Treg) gene signatures, which are known to participate in immunosuppression and be detrimental to ICI efficacy
Multiplex immunofluorescence panel focused on T cell exhaustion
Specimens of 64 mTLS positive NSCLC patients were then analyzed by digital pathology:
- A panel consisting of CD8, PD1, CD39, LAG3, TIGIT, TIM3, and DAPI revealed a high density of intratumoral CD8+PD1+ T cells in the FAP+αSMA+ CAF-High tumors, with a higher expression of exhaustion markers.
- A panel including CD4, CD8, CD20, FoxP3, ICOS, TIGIT and DAPI highlighted an increased presence of CD4+ T cells in the stroma of MYH11+αSMA+ CAF-High tumors, with an increased infiltration of regulatory CD4+FoxP3+ T cells expressing immunosuppressive markers.
Altogether, these findings suggest that the presence of FAP+αSMA+ CAFs and MYH11+αSMA+ CAFs is associated with poor outcomes in mTLS-positive NSCLC patients undergoing ICI treatment. They could serve as biomarkers to stratify patients, and could constitute targets, in combination with strategies addressing T-cell exhaustion, to enhance ICI efficacy in mTLS-positive NSCLC patients.
Interested in the assessment of TLS maturity? Learn about our histopathology services for TLS detection & scoring.
Read paper
Over the past 5 years, Explicyte has supported several cancer immunotherapy programs aimed at inducing or enhancing a specific anti-tumor immune response. Here's a quick overview of our capacities for the advancement of novel cancer vaccines (peptide-based, DC-based, DNA- or RNA-based), cytokine therapies, oncolytic virus-based therapies, immune modulators (such as STING agonists), and immune adjuvants.
We designed a cell-based system to assess the effectiveness of compounds aimed at enhancing the proficiency of antigen-presenting cells (APC), such as dendritic cells (DC), to capture and cross-present the tumor antigens, and the subsequent priming of CD8+ cytotoxic T cells against cancer cells. The assay provides dynamic flexibility, thus fitting different typologies of test compounds, with different modes of action and treatment modalities.
CASE STUDY

Exposure of DCs to a target tumor antigen (TA) peptide enhances their antigenic presentation shown by the increase of its surrogate expression (surface (left) and intracellular (right)) using specific antibodies.

Target TA-exposed DCs lead to optimized priming of CD8 T cells, as shown through the increased IFNγ levels released in CD8 T cell / DC co-cultures.
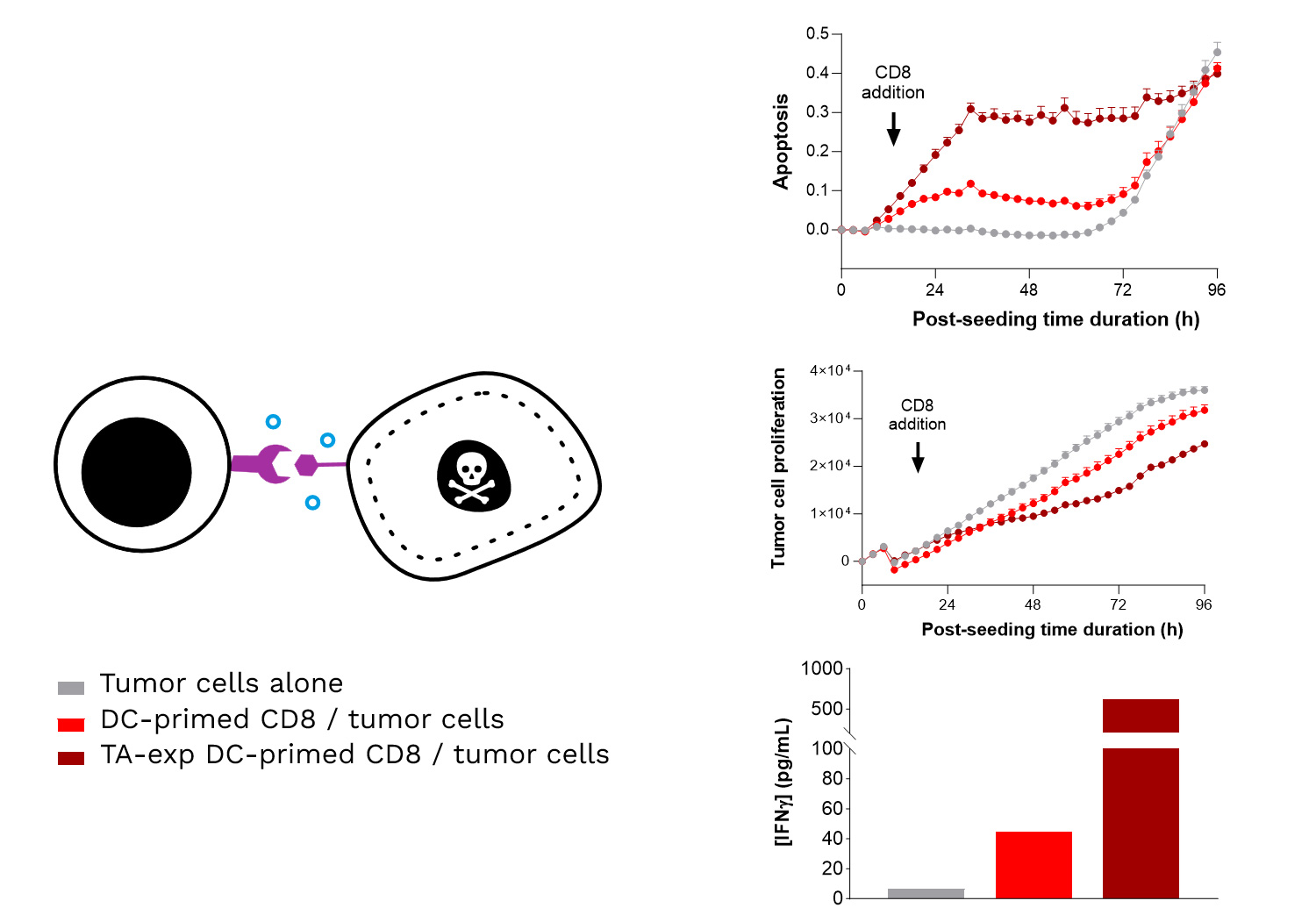
Specific TA-exposed DC-mediated priming of CD8 T cells results in the induction of an adaptive effector T cell-mediated killing towards SK-MEL-5 tumor cells (target TA-positive). Real-time monitoring of SK-MEL-5 tumor cell killing mediated by CD8 T cells, primed by either TA-exposed or unexposed DCs, where apoptosis and tumor cell count were monitored over ~4 days and analyzed. In addition, IFNγ release by CD8 T cells is shown to be increased upon their TA-exposed DC priming, compared to TA-unexposed DCs.
TAKE HOME MESSAGE: Enhancing DC-mediated tumor antigen presentation induces a specific, optimized, adaptive effector T cell-mediated killing response.
A step-by-step overview of our tumor antigen-specific T-cell mediated tumor killing assay:
Starting from FFPE tumor specimens, our translational team can explore and/or validate the expression of target tumor antigens across a set of cancer indications, using various platforms:
Talk to a scientist from our translational team
Specific Tumor Antigen Presentation: In vitro Efficacy & MoA Studies with a dedicated DC-mediated T-cell Killing Assay
We designed a cell-based system to assess the effectiveness of compounds aimed at enhancing the proficiency of antigen-presenting cells (APC), such as dendritic cells (DC), to capture and cross-present the tumor antigens, and the subsequent priming of CD8+ cytotoxic T cells against cancer cells. The assay provides dynamic flexibility, thus fitting different typologies of test compounds, with different modes of action and treatment modalities.
CASE STUDY
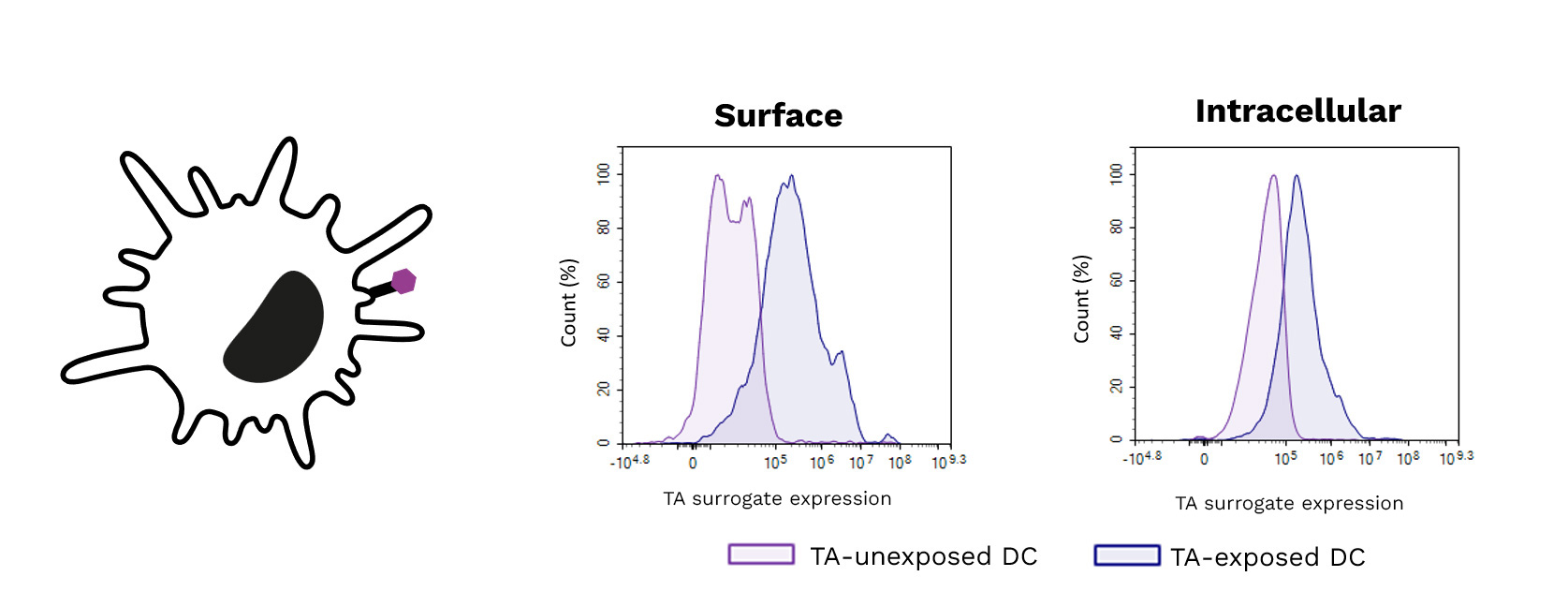
Exposure of DCs to a target tumor antigen (TA) peptide enhances their antigenic presentation shown by the increase of its surrogate expression (surface (left) and intracellular (right)) using specific antibodies.
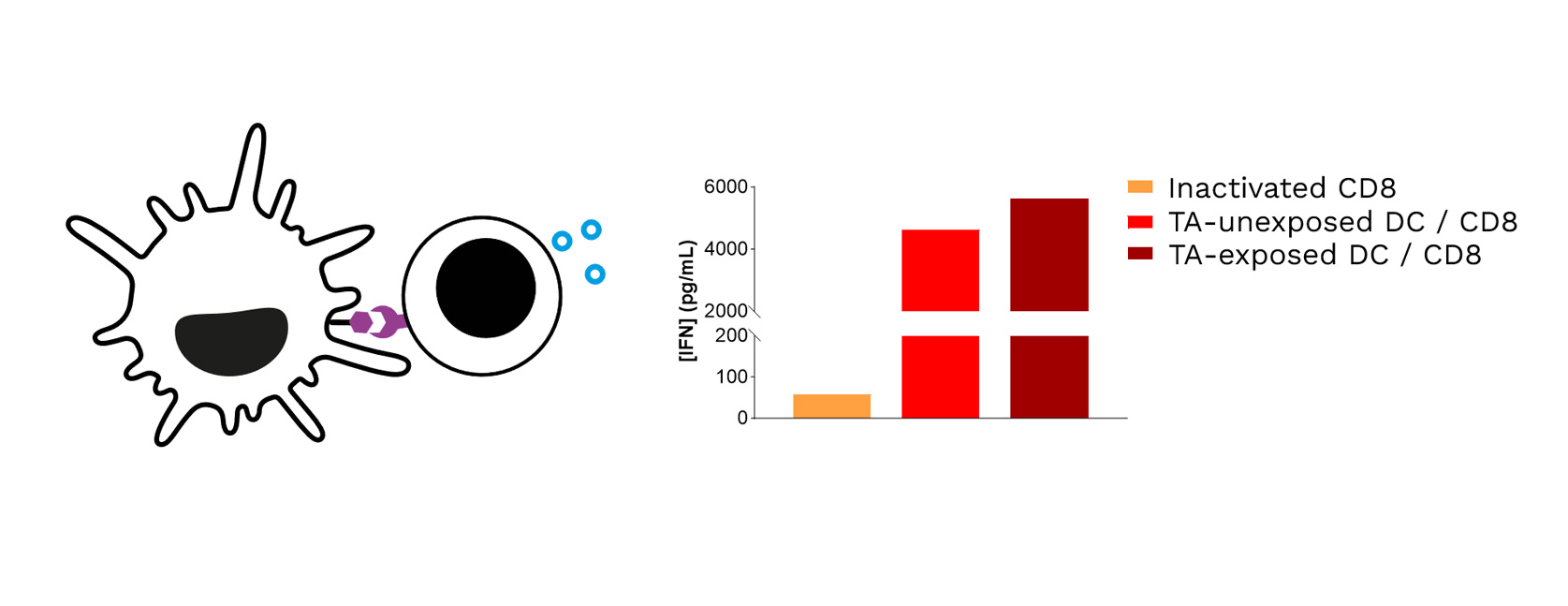
Target TA-exposed DCs lead to optimized priming of CD8 T cells, as shown through the increased IFNγ levels released in CD8 T cell / DC co-cultures.
Specific TA-exposed DC-mediated priming of CD8 T cells results in the induction of an adaptive effector T cell-mediated killing towards SK-MEL-5 tumor cells (target TA-positive). Real-time monitoring of SK-MEL-5 tumor cell killing mediated by CD8 T cells, primed by either TA-exposed or unexposed DCs, where apoptosis and tumor cell count were monitored over ~4 days and analyzed. In addition, IFNγ release by CD8 T cells is shown to be increased upon their TA-exposed DC priming, compared to TA-unexposed DCs.
TAKE HOME MESSAGE: Enhancing DC-mediated tumor antigen presentation induces a specific, optimized, adaptive effector T cell-mediated killing response.
How does it work?
A step-by-step overview of our tumor antigen-specific T-cell mediated tumor killing assay:
- Choice of the right tumor cell lines based on their expression of the target antigens and their representativeness of the cancer indications of interest (>100 in-house human tumor cell lines)
- Isolation of PBMC-originating immune populations required (monocytes, CD8) for autologous co-cultures
- Generation of monocyte-derived APC such as differentiated & mature DCs
- DC - CD8 T cell co-cultures for DC-mediated priming and activation of CD8 cells
- CD8 T cell - target tumor cell co-cultures to capture the tumor-targeted cytotoxic response
- Adequate treatment windows in line with the expected mechanisms of action and eventual promising combination treatments that could hold potential for improving anti-tumor response
- Relevant readouts to capture each cell component (DC cytokines and surface markers, CD8 cytokines and surface markers, target tumor cell apoptosis/proliferation…)
More data ? Some questions ? Contact our team !
Our capacities for the identification & Validation of Target Antigen Expression in Tumor Samples
Starting from FFPE tumor specimens, our translational team can explore and/or validate the expression of target tumor antigens across a set of cancer indications, using various platforms:
- Single-Cell RNA-Seq to identify & quantify tumor antigen expression, and investigate tumor antigen heterogeneity and the immune landscape
- Xenium Single-Cell Transcriptomics to explore tumor antigen patterns and tumor heterogeneity with respect to the immune microenvironment (immune infiltration, response, gene expression profiles and signatures…)
- GeoMx Digital Spatial Profiling for comprehensive antigen mapping within the tumor microenvironment (TME)
- Digital Pathology for precise and multiplexed analysis of antigen expression with the TME - providing a comprehensive view of the tumor antigenic profile and how it could interact with other components of the TME
Talk to a scientist from our translational team
We're excited to share this comprehensive review on Tertiary Lymphoid Structures (TLS), highlighting the work of Florent Peyraud, an oncologist from the Institut Bergonié. Florent completed his PhD thesis at Explicyte under the guidance of Prof. Antoine Italiano (Institut Bergonié/Gustave Roussy). Congratulations, Florent!
This review delves into recent advancements in TLS research, covering:
Our translational team, who co-authored this review, has developed specialized expertise in the detection and scoring of tertiary lymphoid structure (TLS), combining digital pathology and pathologist-assisted image analysis.
Read the article
This review delves into recent advancements in TLS research, covering:
- TLS composition, maturation status, and prevalence across tumor types
- The role of TLS in antitumor immunity and prognosis, including their predictive value in response to immunotherapy
- Therapeutic strategies: drivers of TLS formation, relevant preclinical models, and the induction of TLS using cytokines, chemokines, or existing cancer treatments.
Our translational team, who co-authored this review, has developed specialized expertise in the detection and scoring of tertiary lymphoid structure (TLS), combining digital pathology and pathologist-assisted image analysis.
Read the article
You're developing a small molecule, a biologic (mAb, bispecific, fusion protein), or a vaccination or probiotic-based strategy in immuno-oncology?
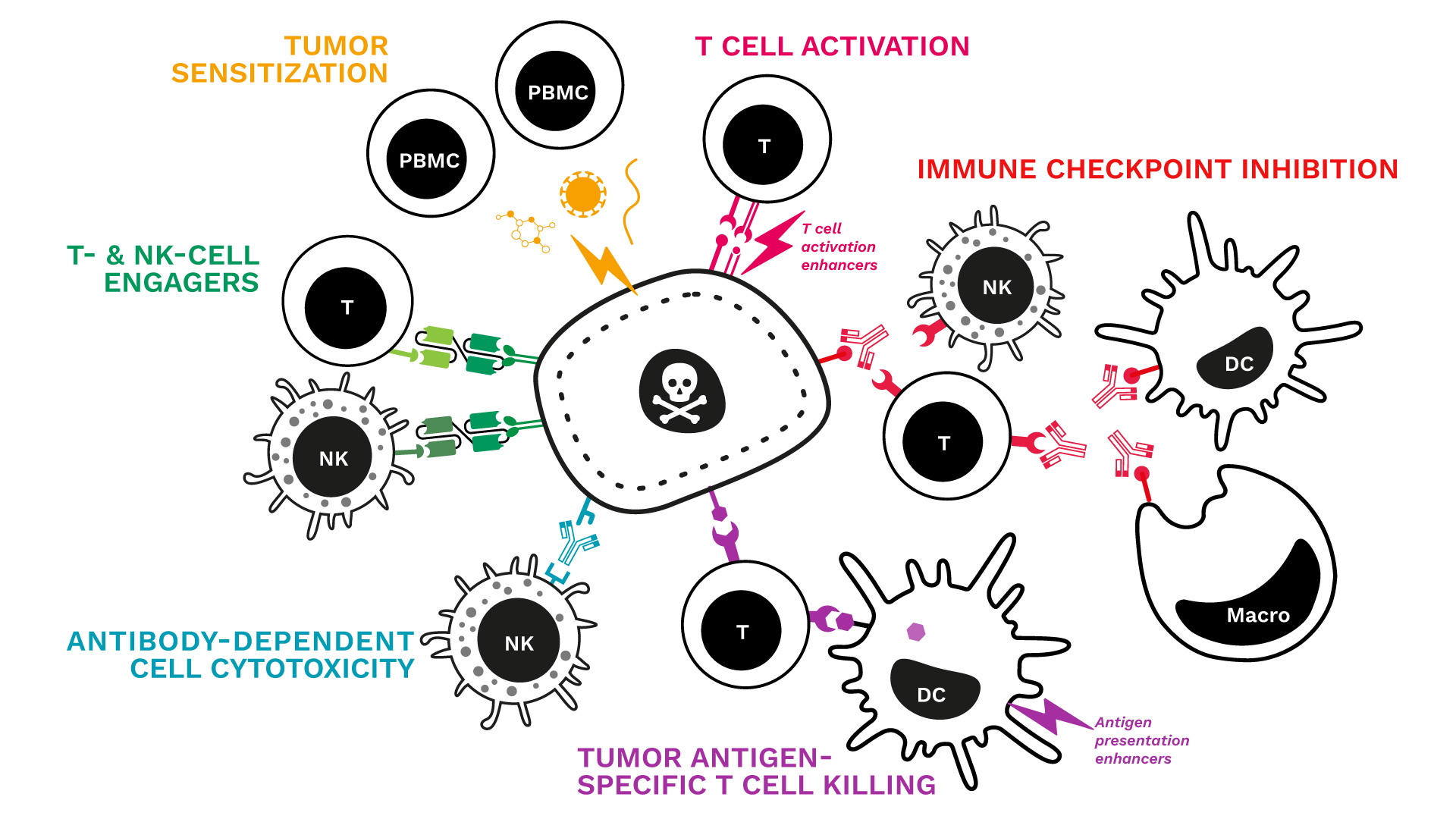
We just updated our range of immune-mediated tumor killing assays!
Key features:
See our specialized killing assays and related data:
We just updated our range of immune-mediated tumor killing assays!
Key features:
- Human co-cultures: the cancer cell line of your choice is cultured with primary immune cells (PBMCs or specific subsets such as Ts or NKs)
- Tumor cell death monitored over time by live-cell imaging
- Immune response captured through the measurement of key cytokines
- A comprehensive analytical platform to further explore and document the profile and mechanism of action of your candidate
See our specialized killing assays and related data:
- Tumor sensitization & killing
- T-cell activation & tumor killing
- Immune checkpoint blockade & tumor killing
- Tumor-antigen specific T cell killing
- Antibody-dependent cell cytotoxicity (NK tumor killing)
A question? A specific request? Contact us !
This retrospective work, led by Prof. Antoine Italiano (Bergonié Comprehensive Cancer Center) and funded by AstraZeneca and the Nouvelle-Aquitaine Regional Council, is based on the BIP study (NCT02534649). It seeks to evaluate the predictive value of CD8+ T cell exhaustion in lung adenocarcinoma patients treated with immune checkpoint inhibitors.
Assessment of TIL Exhaustion by Multiplex Immunohistofluorescence (mIHF)
Explicyte applied a panel indicative of tumor-infiltrating lymphocyte (TIL) exhaustion (CD8, CK7, LAG3, PD1, TIGIT, and TIM3) to tumor samples from 166 patients with advanced lung adenocarcinoma, collected before their treatment with anti-PD1/PD-L1 immunotherapy. Our mIHF analysis revealed that non-responders featured a high proportion of CD8+ T cells co-expressing PD1 and at least one other marker of exhaustion (LAG3, TIGIT, or TIM3). This profile was associated with poorer clinical outcomes, independently of other factors such as age, gender, and PD-L1 status assessed by IHC.
Whole Transcriptome Analysis Reveals a 25-Gene Signature Predictive of CD8+ T Cell Exhaustion and Response to Immunotherapy
Our translational team then analyzed a total of 135 FFPE samples - characterized for their exhaustion status by mIHF - using the HTG transcriptome panel (19,000+ targets). From this analysis, we identified a 25-gene transcriptomic signature strongly associated with the exhaustion phenotype, demonstrating high predictive accuracy. The robustness of this signature was validated in external datasets from NSCLC clinical trials (POPLAR [NCT02517892], OAK [NCT0200822], and MATCH-R [NCT02517892]).
The Predictive Value of the 25-Gene Exhaustion Signature Extends Beyond Lung Cancer
Datasets from the CA209-038 melanoma trial (NCT01621490) and the JAVELIN Renal 101 (NCT0268400601) trial in renal carcinoma confirmed the robustness of the CD8+ T cell exhaustion signature as a biomarker for stratifying patients likely to benefit from immunotherapy.
Assessment of TIL Exhaustion by Multiplex Immunohistofluorescence (mIHF)
Explicyte applied a panel indicative of tumor-infiltrating lymphocyte (TIL) exhaustion (CD8, CK7, LAG3, PD1, TIGIT, and TIM3) to tumor samples from 166 patients with advanced lung adenocarcinoma, collected before their treatment with anti-PD1/PD-L1 immunotherapy. Our mIHF analysis revealed that non-responders featured a high proportion of CD8+ T cells co-expressing PD1 and at least one other marker of exhaustion (LAG3, TIGIT, or TIM3). This profile was associated with poorer clinical outcomes, independently of other factors such as age, gender, and PD-L1 status assessed by IHC.
Whole Transcriptome Analysis Reveals a 25-Gene Signature Predictive of CD8+ T Cell Exhaustion and Response to Immunotherapy
Our translational team then analyzed a total of 135 FFPE samples - characterized for their exhaustion status by mIHF - using the HTG transcriptome panel (19,000+ targets). From this analysis, we identified a 25-gene transcriptomic signature strongly associated with the exhaustion phenotype, demonstrating high predictive accuracy. The robustness of this signature was validated in external datasets from NSCLC clinical trials (POPLAR [NCT02517892], OAK [NCT0200822], and MATCH-R [NCT02517892]).
The Predictive Value of the 25-Gene Exhaustion Signature Extends Beyond Lung Cancer
Datasets from the CA209-038 melanoma trial (NCT01621490) and the JAVELIN Renal 101 (NCT0268400601) trial in renal carcinoma confirmed the robustness of the CD8+ T cell exhaustion signature as a biomarker for stratifying patients likely to benefit from immunotherapy.
Read paper
In this retrospective study led by Pr. Antoine Italiano (Bergonié & Gustave Roussy Comprehensive Cancer Centers) based on the NEOSARCOMICS trial, we investigated whether microenvironment features of undifferentiated pleomorphic sarcomas (UPS) - an aggressive subtype of soft tissue sarcoma - could predict response to neoadjuvant anthracycline-based chemotherapy.
These results suggest that immune infiltration status can serve to stratify UPS patients before surgical resection. Patients with high immune infiltration benefit from a better prognosis from the start and may respond poorly to chemotherapy. However, neoadjuvant therapy may improve the prognosis of immune-low UPS patients.
- Microenvironment features & prognosis without neoadjuvant therapy: We applied a multiplexed immunohistofluorescence (mIHF) panel on tumor samples from 47 patients with UPS who underwent surgical resection. We found that patients with poor survival had fewer infiltrated immune cells (especially CD8+ cells and M1 macrophages), while patients with better overall survival tended to feature high tumor infiltration.
- Microenvironment features & response to neoadjuvant therapy: We then analyzed the gene expression of baseline samples from 24 patients with resectable UPS, who were treated with neoadjuvant chemotherapy. We found that a good response was associated with a high proliferation and low immune infiltration phenotype. This finding was confirmed with the multiplex mIHF panel: patients who poorly respond to neoadjuvant therapy feature high immune infiltration. Plasma proteomics confirmed the upregulation of cell cycle pathways in low immune infiltration patients.
These results suggest that immune infiltration status can serve to stratify UPS patients before surgical resection. Patients with high immune infiltration benefit from a better prognosis from the start and may respond poorly to chemotherapy. However, neoadjuvant therapy may improve the prognosis of immune-low UPS patients.
Read paper in the Journal Hematology & Oncology
Explicyte is the first immuno-oncology CRO in Europe equipped with the 10x Xenium platform for single-cell spatial transcriptomics! Training by the 10x Genomics team started! Why is it exciting for immuno-oncology?
“For our translational and data science team, the Xenium platform opens unexplored avenues for analyzing tumors and their microenvironment in response to various cancer modalities, including immunotherapies.
For our sponsors, gaining a deep understanding of the gene expression profiles and spatial distribution of individual cells within a tumor specimen can provide unique insights into understanding tumor biology, and mechanisms at play in anti-cancer response or resistance to treatment. It creates opportunities to identify new targets and biomarkers to predict clinical outcomes for cancer patients.
As a researcher, I’m eager to watch tumor cells, immune cells, cancer-associated fibroblasts with the Xenium eye, and to witness the single-cell architecture of immune infiltrates and tertiary lymphoid structures. We are truly excited as we enter a new era of cancer discovery."
Alban Bessede, CEO of Explicyte Immuno-Oncology.
About the 10x Xenium platform:
Learn about our services for target identification & biomarker discovery in immuno-oncology.
“For our translational and data science team, the Xenium platform opens unexplored avenues for analyzing tumors and their microenvironment in response to various cancer modalities, including immunotherapies.
For our sponsors, gaining a deep understanding of the gene expression profiles and spatial distribution of individual cells within a tumor specimen can provide unique insights into understanding tumor biology, and mechanisms at play in anti-cancer response or resistance to treatment. It creates opportunities to identify new targets and biomarkers to predict clinical outcomes for cancer patients.
As a researcher, I’m eager to watch tumor cells, immune cells, cancer-associated fibroblasts with the Xenium eye, and to witness the single-cell architecture of immune infiltrates and tertiary lymphoid structures. We are truly excited as we enter a new era of cancer discovery."
Alban Bessede, CEO of Explicyte Immuno-Oncology.
About the 10x Xenium platform:
- Compatible with standard tumor specimens (frozen & FFPE)
- TME architecture is visualized at subcellular level with a 5000-gene panel
- Single-cell analysis & segmentation with AI-driven algorithm
Learn about our services for target identification & biomarker discovery in immuno-oncology.