As a first biomarker assay developed under this quality management system, Explicyte now offers a multiplex immunofluorescence (IF) test to detect and score tertiary lymphoid structures (TLS) in tumor samples, supporting the identification of patients most likely to benefit from cancer immunotherapies.
https://youtu.be/bmokMO6jCHc
A 10-year track record in histopathology for immuno-oncology programs
Building on more than ten years of experience in digital pathology and translational immuno-oncology programs, Explicyte has validated 150+ tumor and immune markers across sponsored studies. The company integrates in-house FFPE tissue processing, automated IHC and IF staining, high-throughput whole-slide imaging, and standardized scoring workflows (AI- and/or pathologist-assisted) to deliver biomarker assays with high reproducibility across cohorts.
Enabling clinical-grade biomarker programs & future companion diagnostics
“Our move into clinical biomarkers is being driven by the rapid expansion of precision oncology. Drug developers increasingly rely on predictive tissue biomarkers to guide patient selection and trial stratification,” said Alban Bessède, PhD, founder and CEO of Explicyte. “While we already had the technological platforms and scientific expertise, ISO certification was essential to support sponsors at clinical stages. ISO 13485 also opens up new perspectives for the development of companion diagnostics for oncology programs.”
First validated clinical biomarker panel: multiplex TLS assessment in tumor tissues
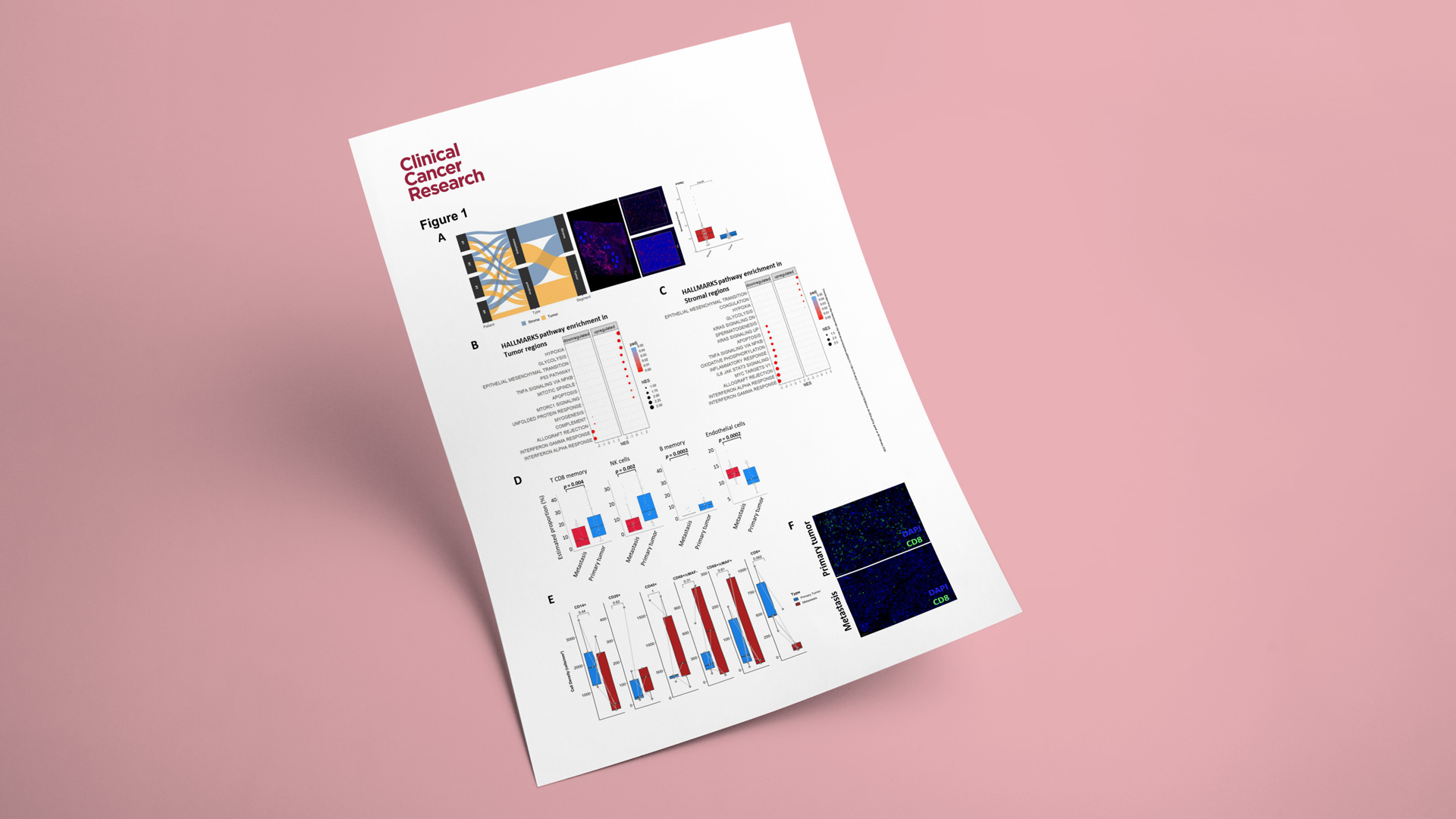
As part of its ISO-certified tissue biomarker offering, Explicyte has developed a multiplex IF panel enabling standardized detection and scoring of TLS in tumor samples. Over the past five years, Explicyte has contributed to several translational studies published in journals such as Nature Cancer and Nature Medicine, which established the clinical relevance of TLS presence and maturity as a predictive biomarker of response to cancer immunotherapies. This validated assay, interpreted by trained pathologists, is now available for routine TLS screening in oncology clinical trials.
Biomarker services to be extended toward single-cell spatial biology
In addition to histopathology, Explicyte continues to invest in advanced translational technologies. In 2025, the company expanded its capabilities with the 10x Genomics suite of single-cell and spatial biology technologies, for which it obtained 10x Genomics’ Certified Service Provider designation.
“Our goal over the next year is to expand our ISO-certified biomarker framework to include single-cell and spatial transcriptomics workflows, as clinical programs increasingly move toward multi-omic, tissue-resolved readouts,” added Dr. Bessède.
About Explicyte's ISO-certified clinical biomarker services
Explicyte is certified ISO 13485:2016 and ISO 9001:2015 by Euro-Quality System (certificates 260133/1637F/1 and 260133/1637F/2) to support precision oncology trials with:
• Design and development of novel tissue biomarkers (single-plex and multiplex IHC/IF panels)
• Tissue biomarker testing services to support therapeutic decision-making
About Explicyte
Explicyte is a contract research organization specializing in precision oncology. Founded in 2015 by immunologist Dr. Alban Bessède, the company has supported over 100 biotech and pharmaceutical partners in the discovery and development of novel therapies for solid tumors. Based at the Institut Bergonié Comprehensive Cancer Center in Bordeaux, Explicyte brings together a multidisciplinary team of 25 scientists, including cell biologists, digital pathologists, medical oncologists, and data scientists. Over the past five years, Explicyte has co-authored 30+ peer-reviewed publications on the molecular mechanisms of response to cancer immunotherapies.
For more information, visit www.explicyte.com
Press contact: Explicyte – Pierre-Emmanuel GAULTIER - pe.gaultier@explicyte.com








![[webinar] Targeting Tregs in Solid Tumors: Anti-CCR8 Therapeutics & Translational Insights Targeting Tregs in Solid Tumors Anti-CCR8 Therapeutics & Translational Insights](https://explicyte.com/wp-content/uploads/2025/10/Targeting-Tregs-in-Solid-Tumors-Anti-CCR8-Therapeutics-Translational-Insights.jpg)